Research Project
On the Challenge of Sustainable Agriculture and Ecosystem under Diverse Environments
Research Team
Lead Researchers:
-
Yoshie Hanzawa, Biology
Collaborators:
- Dr. Eric Deeds (UCLA)
Student Team:
- Dr. Upeksha Hemamali*
- Kimberly Kagan, M.S. student*
- Tanya Pelayo, M.S. student
- Mahwish Zahid, B.S. student
- Silvir Luna, B.S. student
Note: names marked with an asterisk (*) indicate current students
Funding
- Funding Organization:
- Funding Program:
SYNOPSIS
- Sustainable crop production and natural ecosystems under changing environments remain a significant challenge.
- It is important to mitigate the impact of changing environments and to expand crop cultivation to novel environments.
- There is a significant knowledge gap:
- How do specific environmental variables affect plant performance?
- What genetic or epigenetic factors control plant responses to such environmental variables?
Abstract
Sustainable crop production and natural ecosystems have been threatened by changing climate conditions. Addressing this problem requires in-depth knowledge in how specific environmental variables affect plant performance, and what genetic or epigenetic factors control plant responses to such environmental variables. The aim of this study is to gain better understanding of how crop plants perceive and acclimate to environmental variables at the molecular level, with a specific focus on the molecular mechanisms controlling plant’s developmental transition in response to temperature fluctuations. We will characterize the roles of epigenetics and alternative splicing in plant adaptation to diverse temperature conditions using soybeans (Glycine max), a vital legume crop due to its multifaceted significance in industrial applications and global food systems, by clarifying temperature specificity and regulatory functions of histone variants and splicing isoforms in the control of plant development and adaptation. This work will significantly deepen our understanding of plant responses to temperature stimuli in the control of plant development and adaptation, and provide essential knowledge towards improved stress resilience, performance and productivity of crop plants, hence, long-term improvement and sustainability of agriculture and ecosystems.
Motivation/Research Problem
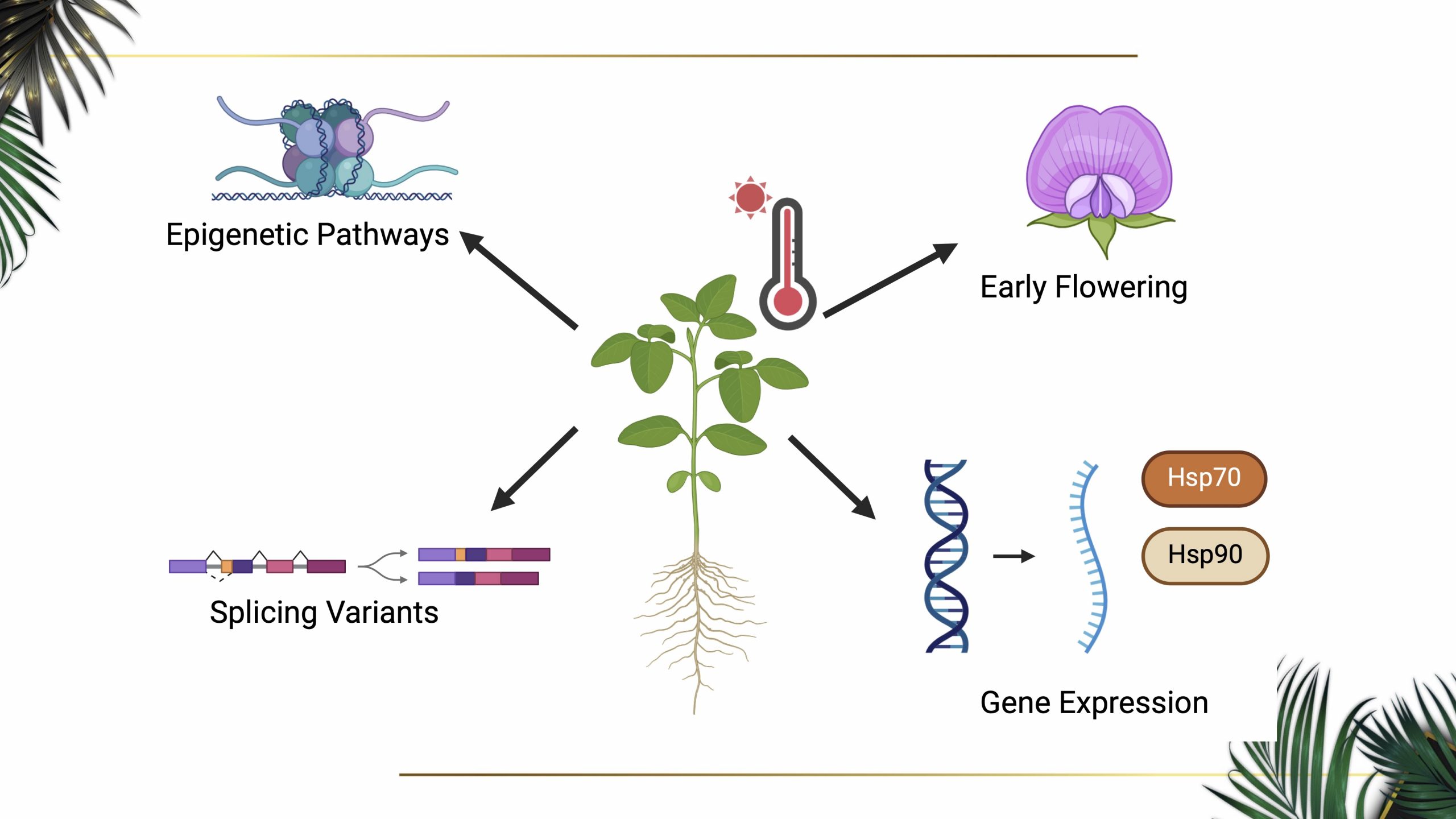

Credits: Faqiang Wu, Dianna Dzheyranyan, Michelle Alcantara
Research Questions and Research Objectives
-
Gain better understanding of how crop plants perceive and acclimate to environmental variables.
-
Clarify the molecular mechanisms controlling plant’s developmental transition in response to temperature fluctuations.
-
Build mathematical models of the gene regulatory network to enable prediction of plant response to diverse environments.
Research Methods
-
Characterizing temperature-specific expression of epigenetic factors and splicing isoforms using RNA-sequencing and quantitative reverse transcription polymerase chain reaction (qRT-PCR).
-
Identifying downstream genes using network inference algorithms.
-
Experimentally validating inferred regulatory networks using CRISPR genome editing and chromatin immunoprecipitation technologies.
-
Develop differential equation models of the gene regulatory networks.
Research Results and Deliverables
-
Identified alternative splicing variants and epigenetic factors that plants may employ in order to respond to temperatures.
-
Obtained inferred regulatory networks involving epigenetic mechanisms and circadian clock genes.
-
Empirically verified several inferred regulatory interactions.
Commercialization and/or Societal Impact Opportunities
-
Application: Prediction of crop performance and engineering climate resilient plants
-
Key Values: Sustainable agriculture and ecosystem
-
Potential Customers: Biotech industry, agriculture and seed company, breeders
Research Timeline
Start Date: August 1, 2024
End Date: July 31, 2026
Lead Researchers:
-
Yoshie Hanzawa, Biology
Collaborators:
- Dr. Eric Deeds (UCLA)
Student Team:
- Dr. Upeksha Hemamali*
- Kimberly Kagan, M.S. student*
- Tanya Pelayo, M.S. student
- Mahwish Zahid, B.S. student
- Silvir Luna, B.S. student
Note: names marked with an asterisk (*) indicate current students
Funding
- Funding Organization:
- Funding Program:
