On the Challenge of Sustainable Agriculture and Ecosystem under Diverse Environments
Research Team
Lead Researchers:
-
Yoshie Hanzawa, Biology
Collaborators:
Student Team:
- Kimberly Kagan, M.S. student
Tanya Pelayo, M.S. student
Mahwish Zahid, B.S. student
Silvir Luna, B.S. student
Funding
- Funding Organization:
- Funding Program:
Abstract
Sustainable crop production and natural ecosystems have been threatened by changing climate conditions. Addressing this problem requires in-depth knowledge in how specific environmental variables affect plant performance, and what genetic or epigenetic factors control plant responses to such environmental variables. The aim of this study is to gain better understanding of how crop plants perceive and acclimate to environmental variables at the molecular level, with a specific focus on the molecular mechanisms controlling plant’s developmental transition in response to temperature fluctuations. We will characterize the roles of epigenetics and alternative splicing in plant adaptation to diverse temperature conditions using soybeans (Glycine max), a vital legume crop due to its multifaceted significance in industrial applications and global food systems, by clarifying temperature specificity and regulatory functions of histone variants and splicing isoforms in the control of plant development and adaptation. This work will significantly deepen our understanding of plant responses to temperature stimuli in the control of plant development and adaptation, and provide essential knowledge towards improved stress resilience, performance and productivity of crop plants, hence, long-term improvement and sustainability of agriculture and ecosystems.
Motivation/Research Problem
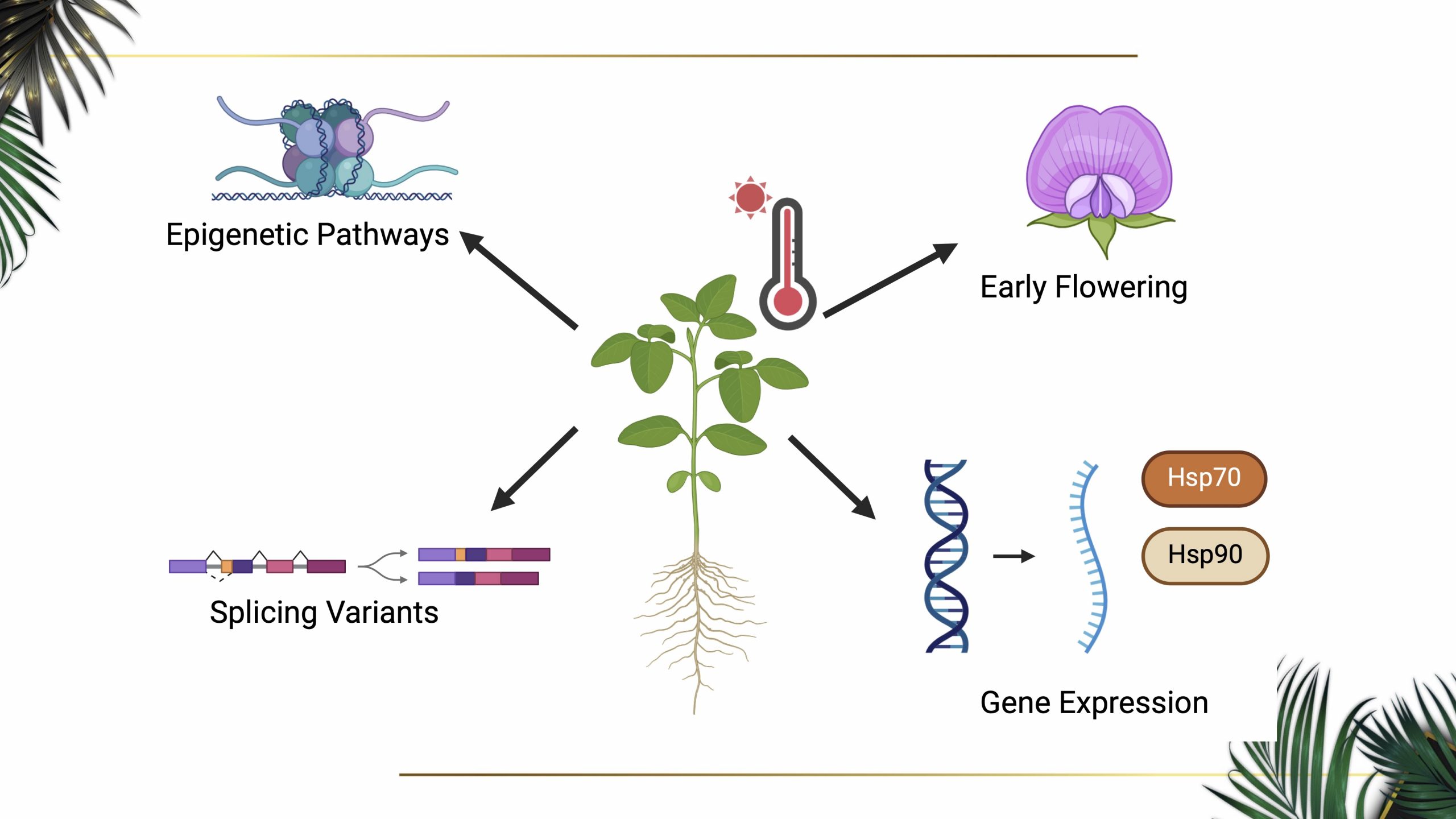
Research Questions and Research Objectives
Our aim in this study is to gain better understanding of how crop plants perceive and acclimate to environmental variables at the molecular level, with a specific focus on the molecular mechanisms controlling plant’s developmental transition in response to temperature fluctuations. Plants adapt to high-temperature environments by adjusting the timing of reproductive growth: some species accelerate reproductive growth to expedite offspring production, while others delay the process to mitigate the adverse effects of heat on reproductive development. Cold stress may inhibit reproductive transition and cause damages or disruption to metabolic processes, leading to reduced growth rates and potentially trigger dormancy or cold acclimation in some plant species.
Our recent work with soybeans has identified two potential mechanisms that plants may employ in order to respond to temperatures: epigenetics and alternative splicing. Epigenetics, organisms’ ability to change their characteristics without altering their DNA sequences, plays a crucial role in plant development as it allows plants to respond to environmental cues. Since epigenetics does not require permanent genetic modifications, this process can occur rapidly under changing environments. The mechanisms of epigenetic effects involve chemical modifications such as methylation and acetylation that occur on DNA nucleotides and on histone proteins attached to DNA. Recent studies suggest that certain DNA or histone modifications and specific histone variants can influence transcriptional activities of the genome and control plant thermosensory mechanisms, including plant’s developmental transition and reproductive growth. Alternative splicing is a post-transcriptional gene regulation mechanism unique to eukaryotic organisms that facilitates the production of multiple transcript and protein isoforms from a single gene. This process is frequently observed in response to environmental abiotic stressors, allowing organisms to adapt and respond to changing conditions. Alternative splicing involves the selective inclusion or exclusion of exons in a precursor messenger RNA (mRNA) molecule. This process generates a diverse array of variably spliced mRNA transcripts, which serve as templates for protein translation. The resulting protein isoforms may exhibit distinct sequences and possess unique functional properties in plant adaptation to specific environmental conditions.
In this project, we will characterize the roles of histone variants and splicing isoforms that we have identified previously in soybeans in plant adaptation to diverse temperature conditions using CRISPR genome editing and chromatin immunoprecipitation technologies.
Research Methods
To better understand how crop plants perceive and acclimate to environmental variables at the molecular level, we will characterize the roles of epigenetics and alternative splicing in plant adaptation to diverse temperature conditions using soybeans (Glycine max), a vital legume crop due to its multifaceted significance in industrial applications and global food systems. Targeting histone variants and splicing isoforms that we have identified previously, we will clarify their temperature specificity and regulatory functions in the control of plant development and adaptation. Our approaches are:
(1) Characterizing temperature-specific expression of histone variants and splicing isoforms using RNA-sequencing and quantitative reverse transcription polymerase chain reaction (qRT-PCR).
(2) Identifying downstream target genes of the temperature-specific histone variants and splicing isoforms using network inference algorithms.
(3) Experimentally validating inferred regulatory roles of the temperature-specific histone variants and splicing isoforms using CRISPR genome editing and chromatin immunoprecipitation technologies.
Research Results and Deliverables
This work will significantly deepen our knowledge of plant responses to temperature stimuli in the control of plant development and adaptation, and provide essential knowledge towards improved stress resilience, performance and productivity of crop plants, hence, long-term improvement and sustainability of agriculture and ecosystems.
Research Timeline
End Date: July 31, 2026
Lead Researchers:
-
Yoshie Hanzawa, Biology
Collaborators:
Student Team:
- Kimberly Kagan, M.S. student
- Tanya Pelayo, M.S. student
- Mahwish Zahid, B.S. student
- Silvir Luna, B.S. student
Funding
- Funding Organization:
- Funding Program:
